-
肝细胞肝癌(hepatocellular carcinoma, HCC)是原发性肝癌中最常见的类型,约占75%~80%。2018年全球的新发病例约84.1万,居恶性肿瘤第六位,约78.2万死亡病例,居恶性肿瘤的第四位,发病率和死亡率有逐年增长的趋势[1]。在中国,肝细胞肝癌发病率和死亡率分别位居恶性肿瘤的第四位和第三位,恶性程度极高[2]。目前,临床上肝癌的治疗手段以外科手术为主,辅以介入治疗、放化疗及靶向药、免疫治疗的多学科综合治疗。尽管治疗方式众多,由于复发与转移等因素的影响,肝癌患者的预后较差,5年生存率低于18%[3-4]。在临床诊疗过程中,TNM分期是评估患者预后情况的经典方法,但由于只能从宏观层面对患者预后进行分析,有一定的局限性。随着医疗水平的提高,基因检测、分子靶向治疗等深入临床,分子水平的预后评判方法是目前的一个研究热点[5]。
转移是一个涉及多步骤的复杂生物学过程,是癌细胞从原发部位向外扩散、侵袭的过程,具有直接浸润、淋巴道转移及血行转移等形式[6]。文献[7-8]发现,转移是肝细胞肝癌的主要生物学特征之一,也是其预后不良的主要原因。近年来,随着高通量测序以及生物信息技术的发展,已有越来越多的肝癌预后模型被建立,以协助判断HCC患者的预后[9-11],但暂时还没有关于转移相关基因的预后模型的报道。因此,本文基于转移相关基因数据集,结合肝细胞癌(the cancer genome atlas, TCGA)的转录组数据和临床数据,构建关于转移相关基因的预后预测模型,并验证该模型的准确性和特异性,以在HCC的预后预测中发挥作用,对肝癌的临床诊疗工作有一定的指导作用。
-
从TCGA数据库下载了374例HCC肿瘤样本和50例癌旁样本,ICGC数据库下载了 243例HCC肿瘤样本和202例癌旁样本,临床信息分别包含年龄、性别、病理分期及患者的生存时间和生存状态。随后,将临床信息不全及生存时间小于30天的HCC样本删除。从人类癌症转移数据库(the human cancer metastasis, HCMDB)下载了转移相关的基因[12],其中有1905个基因在TCGA数据集有表达值,用于风险模型的进一步构建。
-
利用“Wilcoxon”秩和检验对转移相关的HCC表达矩阵进行差异表达分析,设定筛选标准为|log2 (FC)| > 2,FDR < 0.05。采用“pheatmap”包绘制了差异表达的火山图和热图。使用“clusterProfiler”包对差异表达的转移相关基因进行基因本体(gene ontology, GO)功能注释和京都基因和基因组百科全书(Kyoto encyclopedia of genes and genomes, KEGG)通路富集分析,设定矫正后P < 0.05作为富集条件。进行蛋白质互作网络构建,并利用Cytoscape软件可视化互作网络。最后,通过网络节点基因筛选TCGA数据集和ICGC数据集共同的互作基因。
-
首先,结合HCC-TCGA样本的生存时间和生存状态,利用单因素Cox分析方法和Kaplan-Meier筛选有预后意义的转移相关基因。随后,为降低模型的拟合度,使用Lasso回归对单因素结果进行筛选。最后,将Lasso回归结果进行随机分组,分为TCGA训练集(n=172)和TCGA验证集(n=171),采用多因素Cox分析对TCGA训练集构建了预后模型。随后,按照风险评分公式计算每位患者的风险评分,根据患者风险评分的中位值,将TCGA训练集和验证集、ICGC验证集的患者分为高风险组和低风险组。采用“survival”“survminer”包及“time ROC”软件包,分别绘制模型的预后生存曲线、风险评分曲线及时间依赖性受试者工作曲线(receiver operator characteristic, ROC)以评估模型的准确性和特异性。结合患者的临床信息,采用单因素和多因素Cox分析各数据集的风险值是否可作为HCC的独立预后因子。最后,采用人类蛋白图谱数据库(the human protein atlas, HPA)[13]对模型基因的临床组织表达水平进行了验证。
-
采用R软件进行统计分析和图形绘制。差异分析采用“Wilcoxon”秩和检验,模型构建采用Cox分析和Lasso回归分析,生存分析采用Kaplan-Meier分析,P < 0.05为差异具有统计学意义。
-
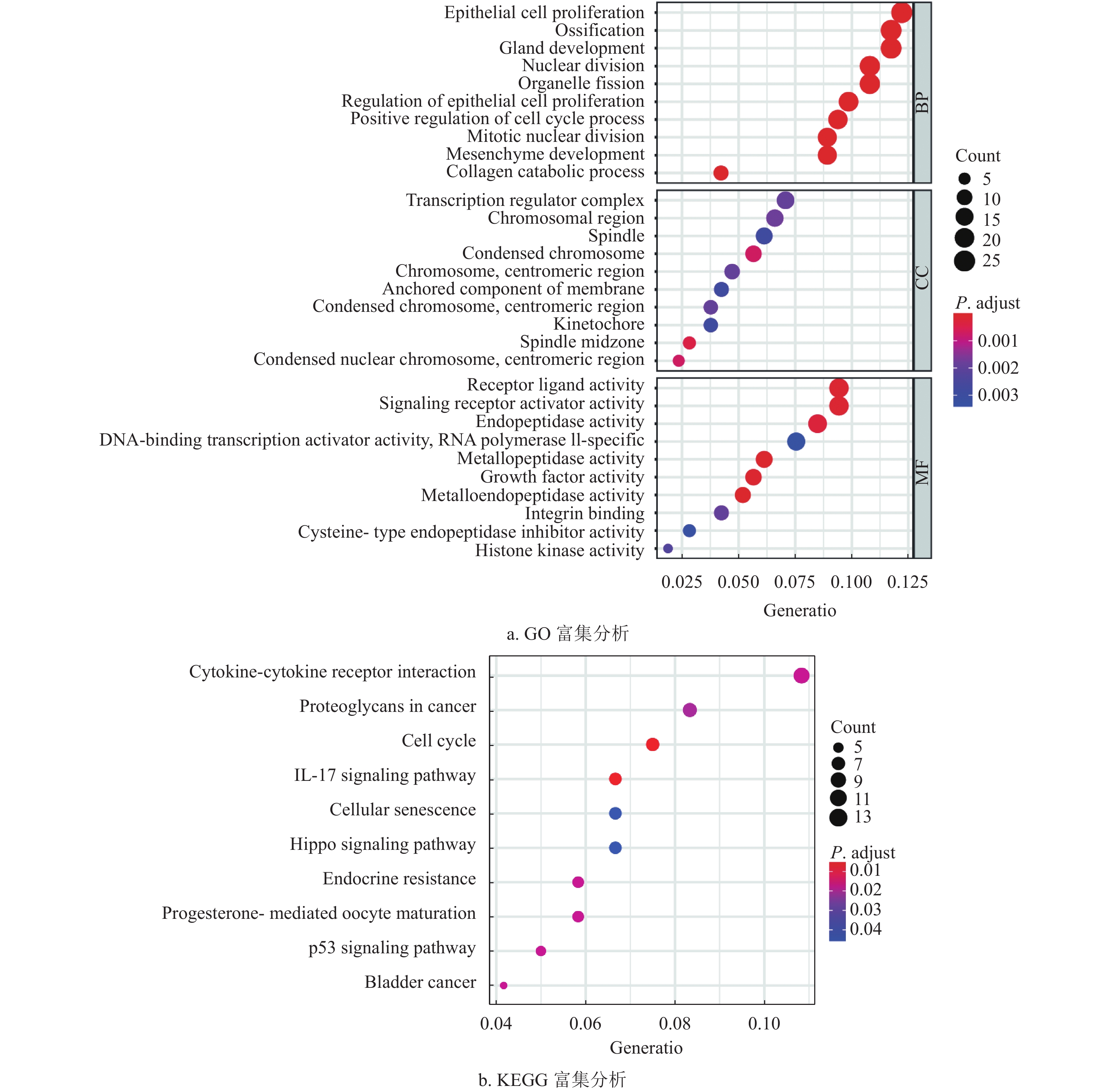
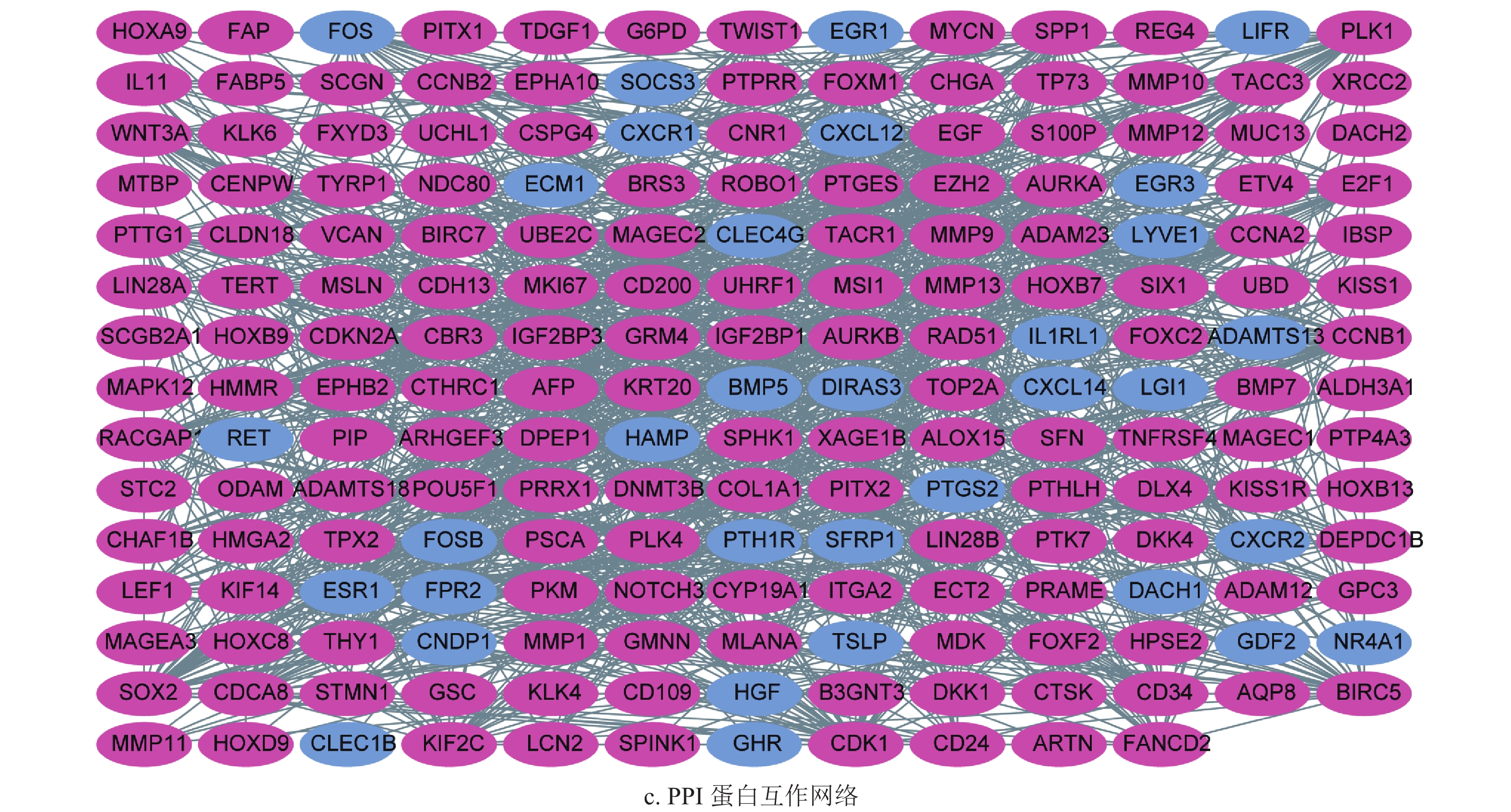
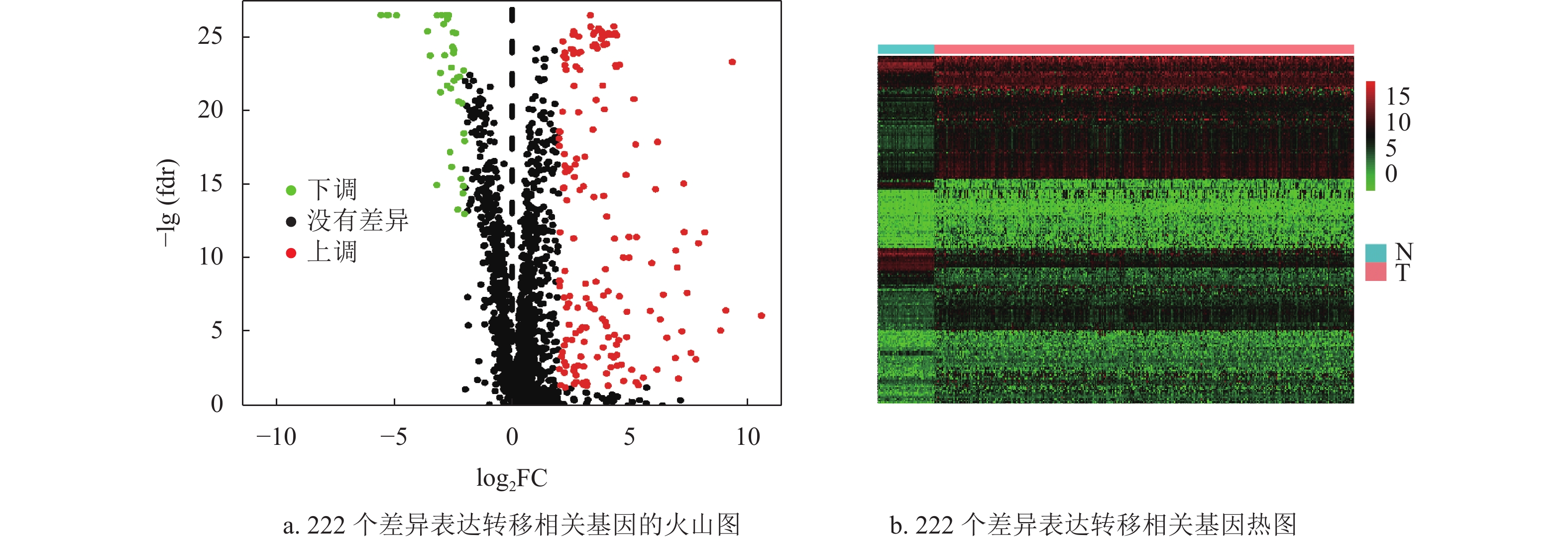
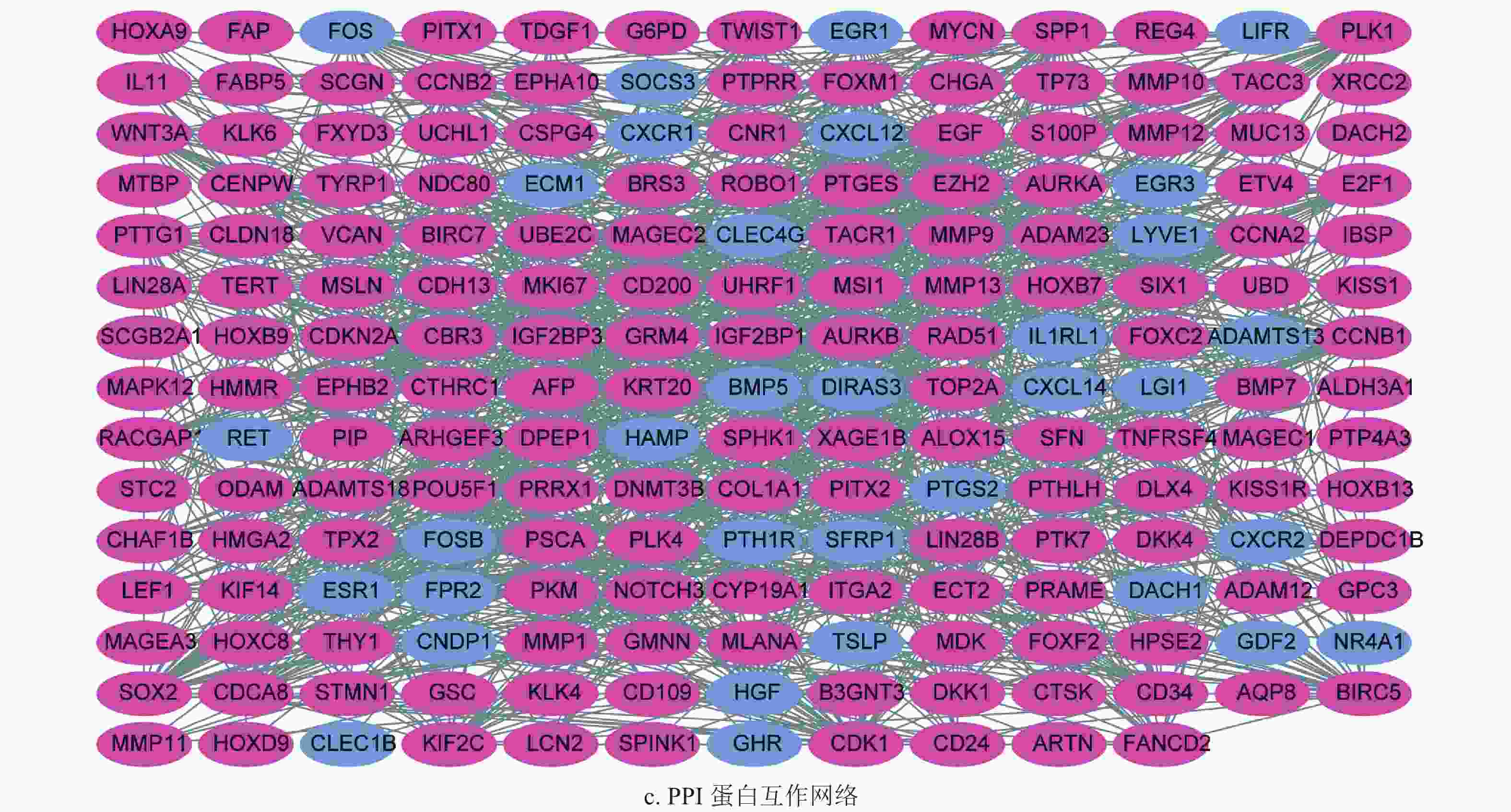
通过差异表达分析,获得了222个差异表达基因,其中39个上调基因,182个下调基因,如图1a和1b所示。将筛选获得的差异基因进行GO功能注释和KEGG通路富集分析,发现差异表达的转移基因主要参与调节上皮细胞增生,胶原分解代谢以及间质发育等生物学过程,其产物主要参与染色体固缩,转录调节复合体等细胞组分,发挥信号转导受体激活和生长因子激活等生物学分子功能,如图2a所示。KEGG通路富集分析表明差异表达的转移相关基因主要参与了细胞因子−细胞因子受体相互作用通路,IL-17和Hippo信号通路,如图2b所示。随后,为了进一步了解差异基因的相互关系,构建了蛋白质互作网络(protein-protein interaction networks, PPI)。通过分析,获得蛋白质互作网络,删除了部分单个存在的基因节点。采用Cytoscape软件进行蛋白质网络的可视化,共获得了194个节点基因,其中红色表示上调基因,蓝色表示下调基因,如图2c所示。其中Generatio是指富集到这个GO条目的基因数目比上所有富集分析的基因数目。
-
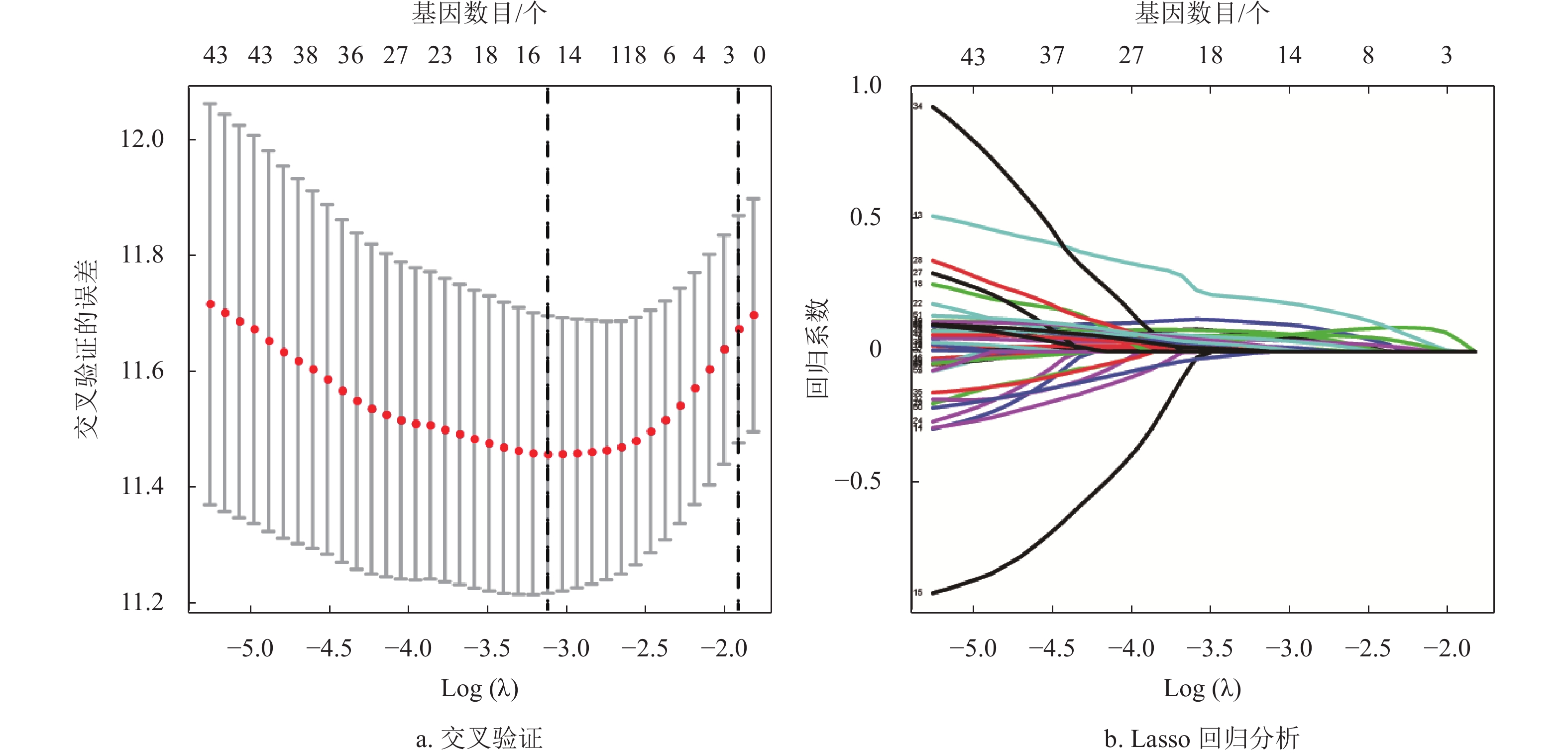
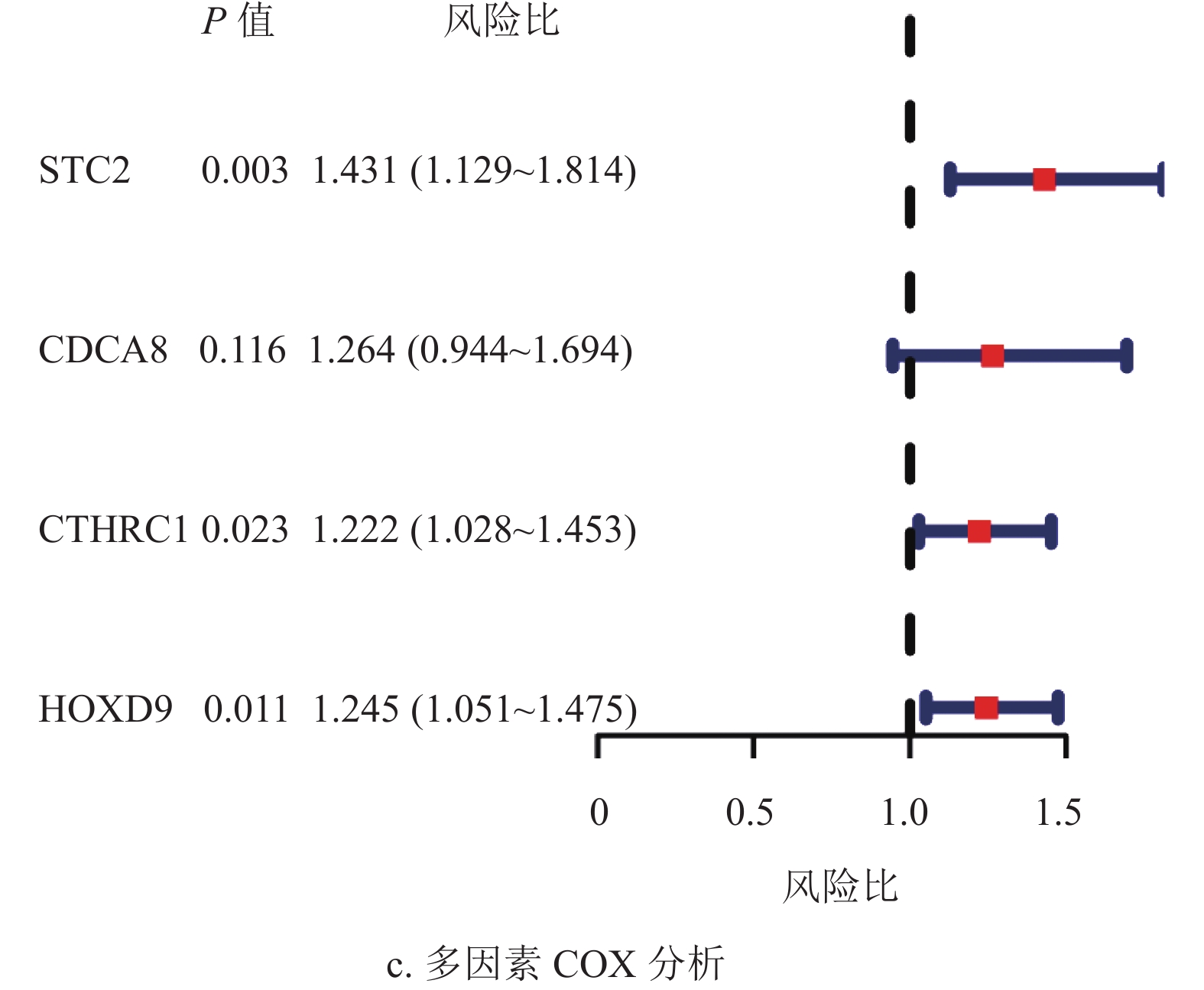
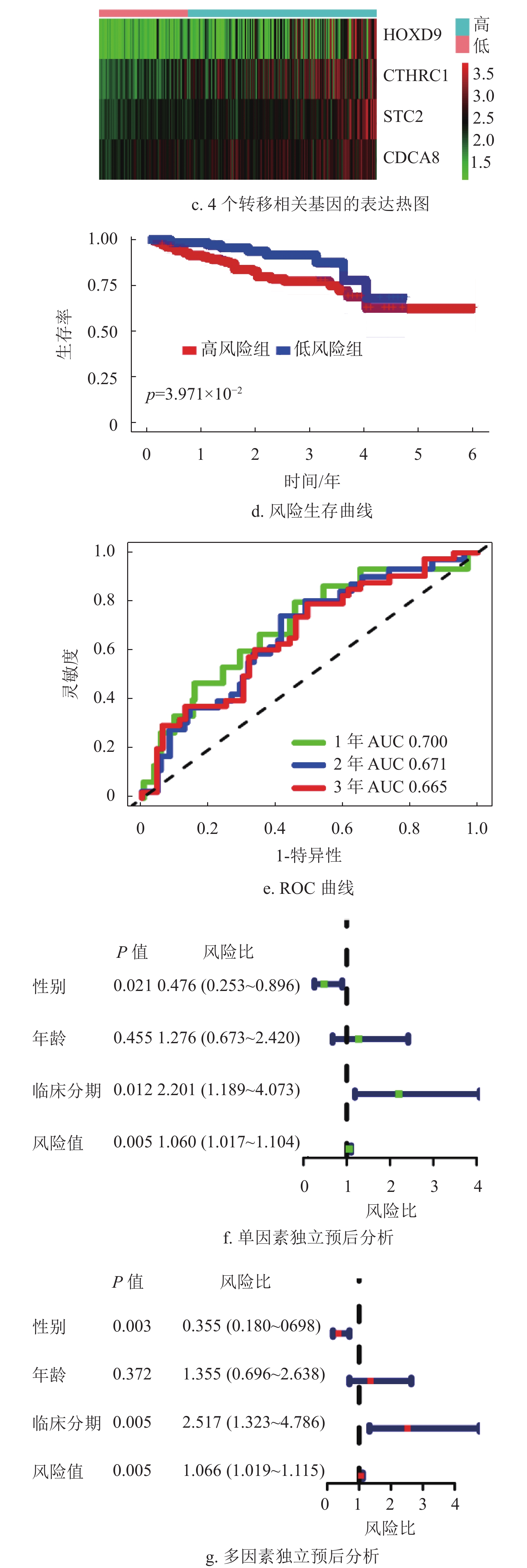
基于蛋白质互作网络的节点基因,采用单因素Cox和Kaplan-Meier分析获得了53个预后相关的转移基因,如表1所示。为降低模型的拟合度,采用Lasso回归分析对预后相关的转移基因进一步筛选,如图3a和3b所示,将筛选结果随机分组。TCGA训练集纳入多因素Cox分析,构建了包含4个转移相关基因的多基因预后模型,如图3c所示。
-
根据模型公式分别计算TCGA训练集,TCGA验证集及ICGC验证集的风险评分,风险评分=0.36×STC2表达值+0.23×CDCA8表达值+0.20×CTHRC1表达值+0.22×HOXD9表达值。
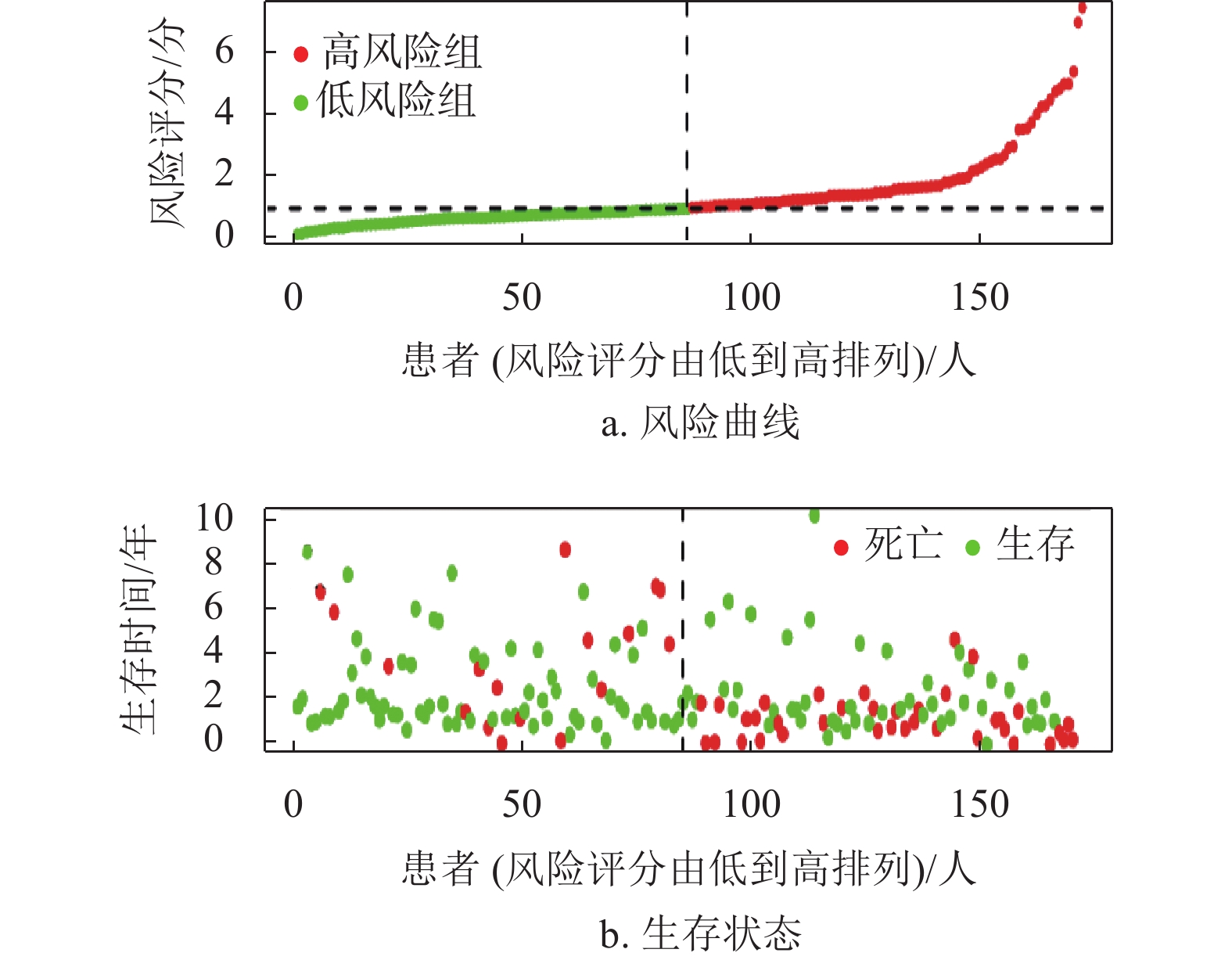
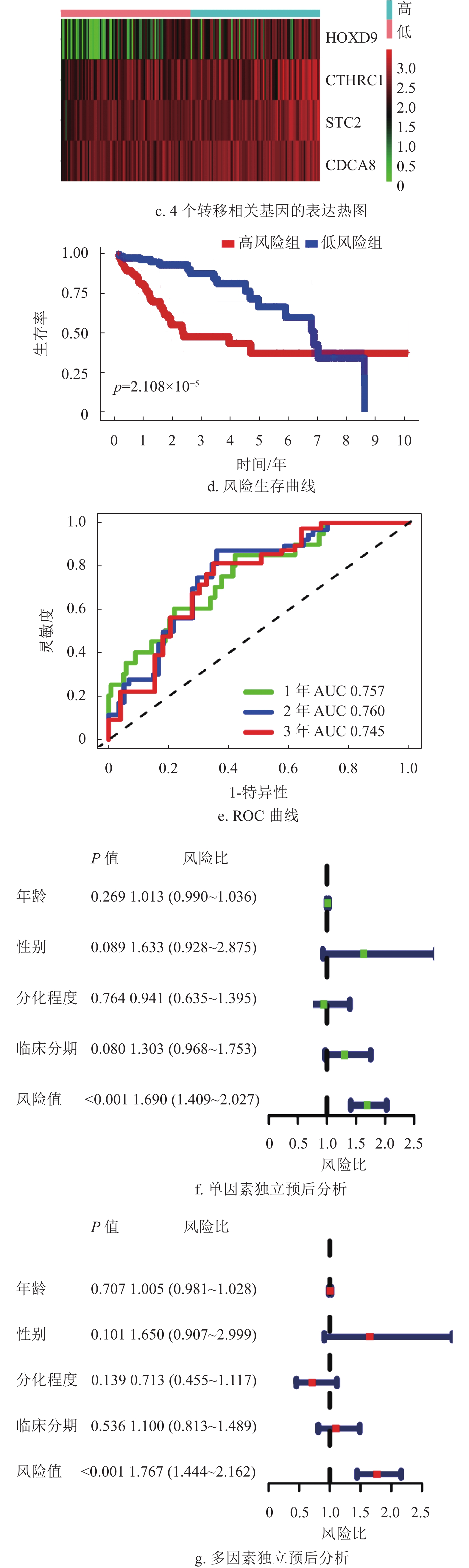
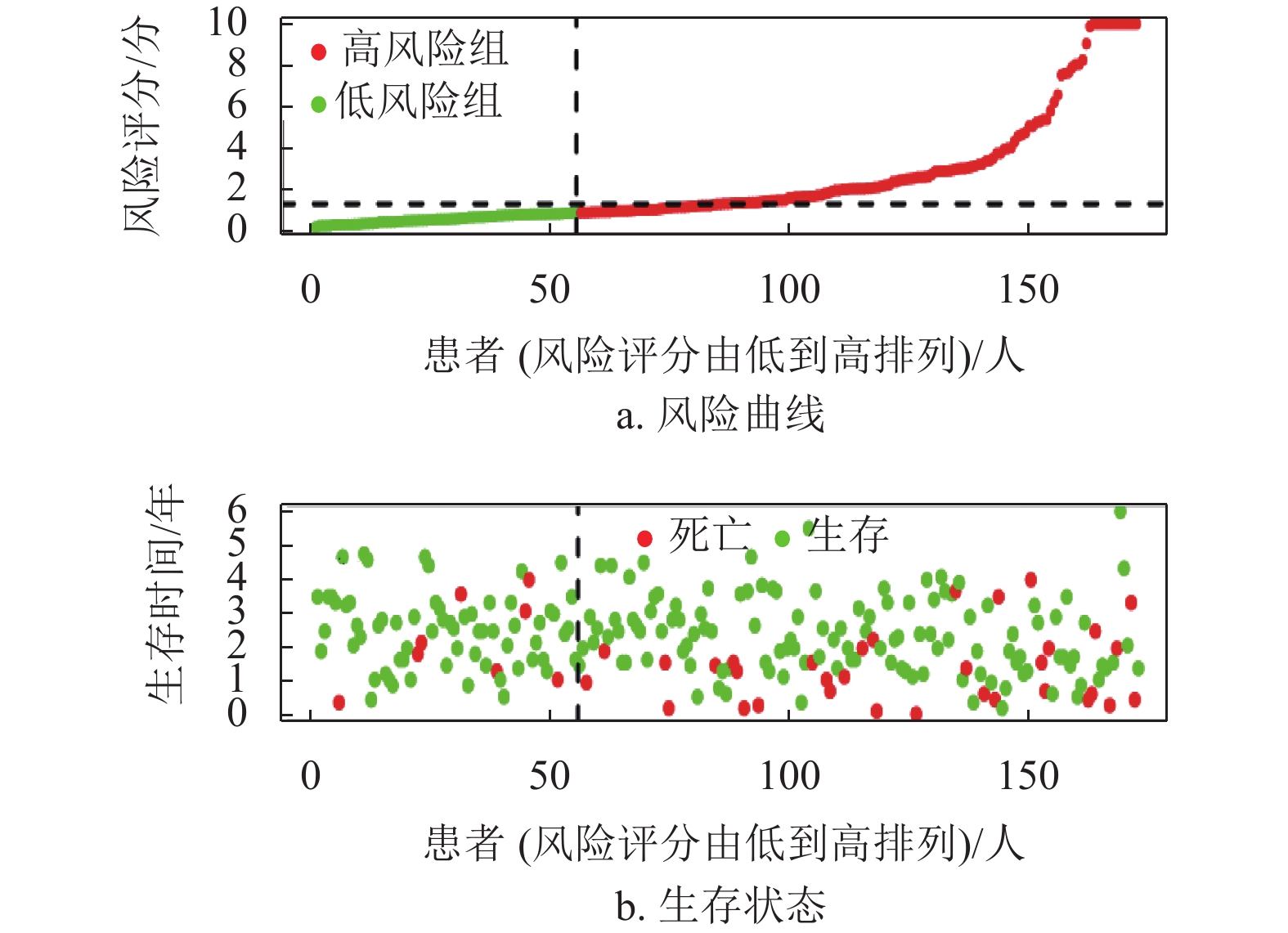
基因名称 风险比 风险比95%置信区间上限 风险比95%置信区间下限 P ADAM12 1.244 1.099 1.407 0.001 KRT20 1.109 1.030 1.194 0.006 CHGA 1.212 1.100 1.335 P < 0.001 STC2 1.405 1.187 1.663 P < 0.001 FOXM1 1.320 1.120 1.556 0.001 SPP1 1.146 1.081 1.214 P < 0.001 CD24 1.137 1.040 1.242 0.005 MAGEA3 1.090 1.030 1.153 0.003 ESR1 0.809 0.723 0.905 P < 0.001 G6PD 1.474 1.287 1.689 P < 0.001 CCNB1 1.425 1.198 1.696 P < 0.001 KIF14 1.316 1.093 1.585 0.004 CDCA8 1.623 1.331 1.980 P < 0.001 CHAF1B 1.279 1.068 1.531 0.007 FANCD2 1.323 1.073 1.631 0.009 LIN28B 1.116 1.038 1.201 0.003 DEPDC1B 1.303 1.126 1.509 P < 0.001 EZH2 1.547 1.224 1.956 P < 0.001 STMN1 1.394 1.175 1.654 P < 0.001 UHRF1 1.315 1.114 1.553 0.001 MKI67 1.372 1.158 1.626 P < 0.001 ECT2 1.456 1.206 1.757 P < 0.001 RACGAP1 1.502 1.201 1.880 P < 0.001 TOP2A 1.275 1.107 1.469 0.001 KIF2C 1.492 1.258 1.770 P < 0.001 CCNA2 1.304 1.116 1.523 0.001 NDC80 1.483 1.216 1.808 P < 0.001 HMMR 1.347 1.127 1.609 0.001 PTTG1 1.270 1.098 1.469 0.001 UBE2C 1.300 1.128 1.498 P < 0.001 PLK1 1.440 1.210 1.714 P < 0.001 BIRC5 1.308 1.128 1.517 P < 0.001 CDK1 1.341 1.136 1.584 0.001 TPX2 1.526 1.274 1.829 P < 0.001 TACC3 1.414 1.161 1.722 0.001 CYP19A1 1.167 1.074 1.267 P < 0.001 SFN 1.140 1.044 1.244 0.004 UCHL1 1.129 1.039 1.226 0.004 CLDN18 1.175 1.049 1.316 0.005 CLEC1B 0.805 0.713 0.909 0.001 MMP12 1.161 1.063 1.267 0.001 CTHRC1 1.237 1.098 1.393 0.001 MMP1 1.333 1.182 1.504 P < 0.001 PITX2 1.179 1.069 1.300 0.001 MMP10 1.266 1.135 1.412 P < 0.001 KLK6 1.226 1.067 1.409 0.004 ETV4 1.159 1.052 1.276 0.003 GHR 0.782 0.699 0.874 P < 0.001 FOSB 0.866 0.778 0.965 0.009 PTPRR 0.766 0.639 0.919 0.004 HOXD9 1.162 1.045 1.291 0.005 XRCC2 1.317 1.086 1.598 0.005 IL1RL1 0.799 0.700 0.912 0.001 在R语言环境下,本文首先分析了TCGA训练集的风险曲线,生存状态以及4个转移相关基因的表达热图如图4a~4c所示。结果表明随着患者的风险评分增加,患者的死亡人数也增加。生存分析结果表明低风险组的患者5年总体生存率比高风险组患者高,如图4d所示,同时采用ROC曲线进一步分析了该模型的特异性和敏感性,该模型1年、2年和3年的AUC值分别为0.757、0.760和0.745,表明该模型能对肝癌患者的生存状态进行一定的预测,如图4e所示。此外,结合HCC患者的临床信息,评估该模型是否能作为预测HCC患者预后的独立因素。单因素和多因素Cox分析结果均表明该模型的风险评分能独立于患者的临床特征而影响HCC患者的预后,如图4f~4g所示。
-
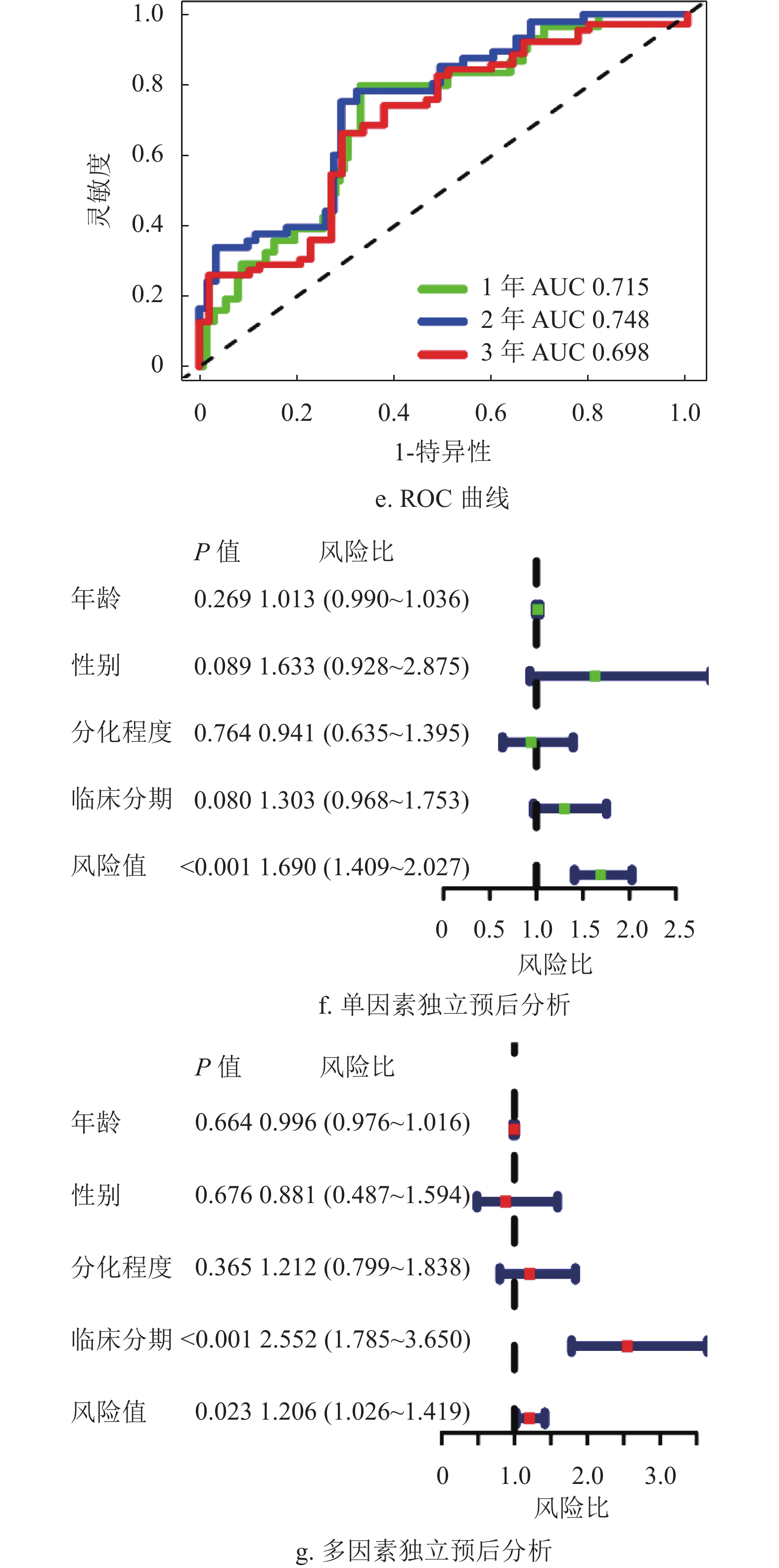
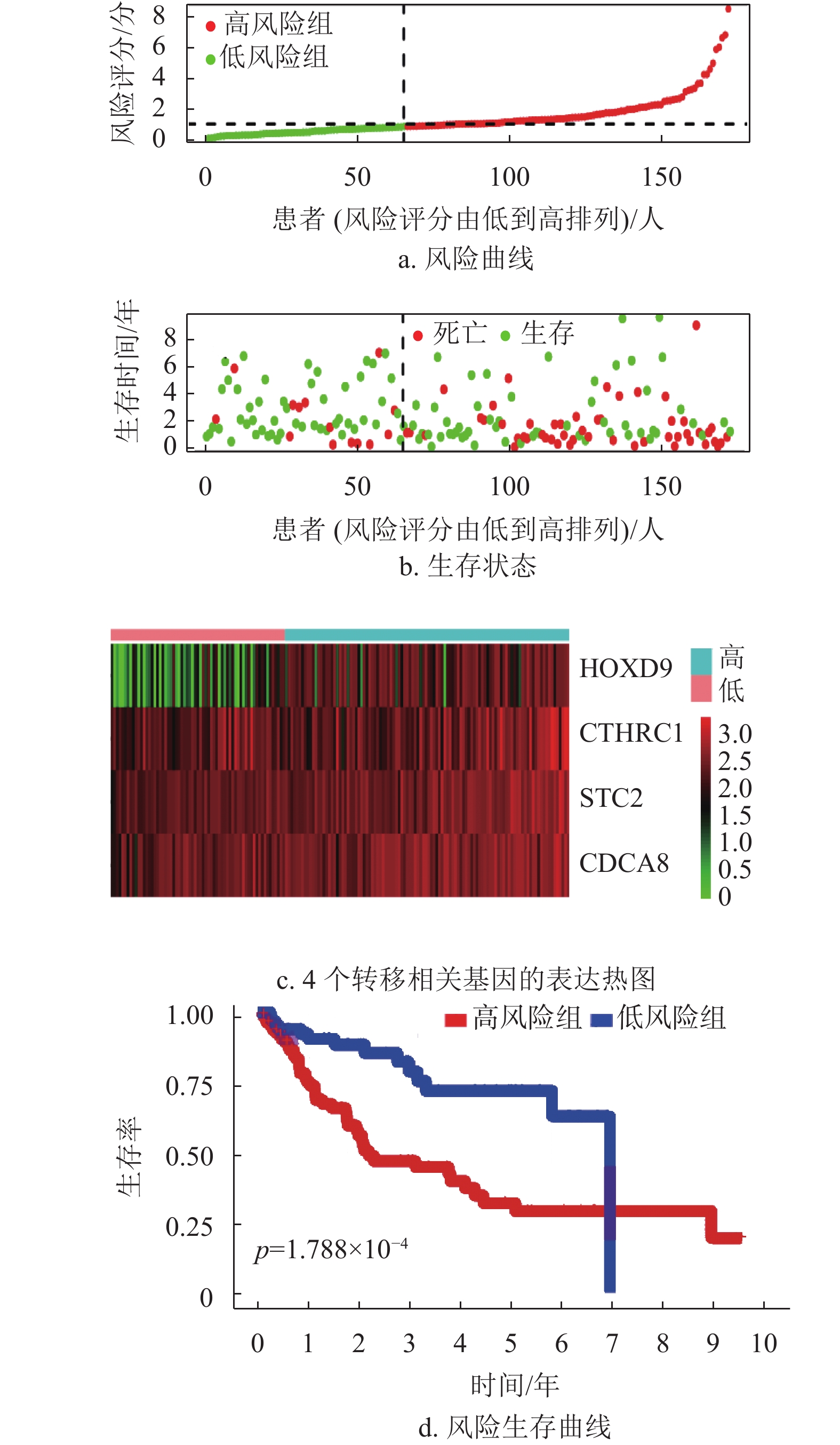
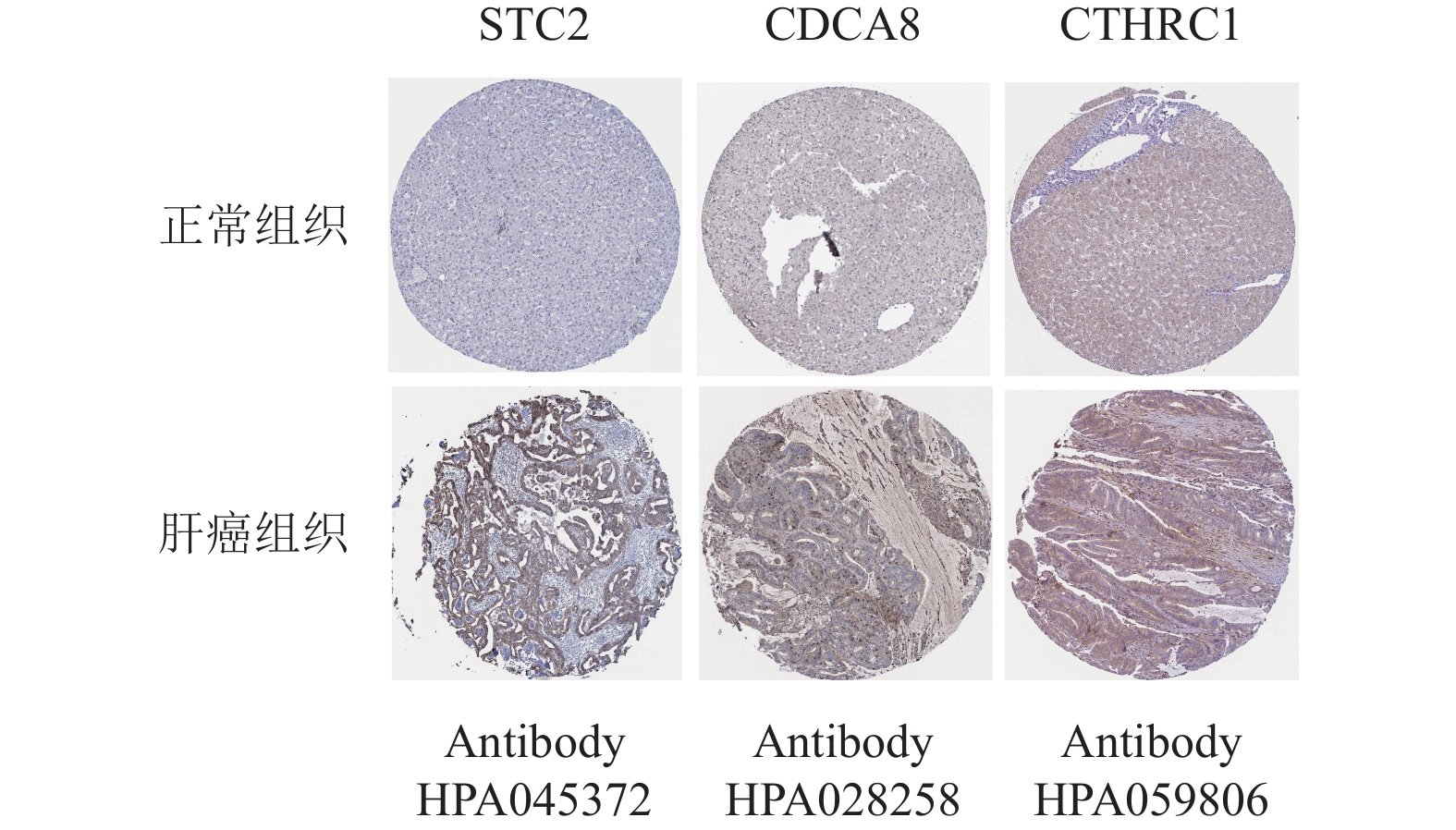
为了进一步验证该模型的准确性和特异性,在TCGA验证集(n =171)和ICGC数据集(n =230)分别再次进行了验证。结果表明,TCGA验证集的结果与TCGA训练集的结果一致,随着风险评分增加,患者的生存越差,低风险组的患者拥有更好的生存,1年、2年和3年的ROC曲线的AUC值分别为0.715、0.748和0.698,再次验证了该模型的特异性和敏感性。同时独立预后分析结果也表明该模型可以作为HCC患者的独立预后因素,如图5所示。此外,通过分析外部验证集ICGC,本文也获得了与TCGA训练集和验证集相同的结果,如图6所示。结合HPA数据库发现,STC2、CDCA8及CTHRC1在肝癌患者临床组织中高表达,在癌旁组织中低表达,如图7所示。同时,据文献[14]的研究,HOXD9基因在肝癌组织中高表达,癌旁组织中低表达。
-
近年来,随着高通量测序技术的发展和公共数据库的开放,可通过定量检测分子预后标记以预测肿瘤的进展。文献[15]研究发现转移相关基因的预后模型在预测胰腺癌患者的预后方面有一定优势。因此,通过整合多个转移相关基因来构建转移相关基因预后模型对HCC患者的预后进行判断,将有助于指导临床决策。
本文采用TCGA和ICGC数据库中的HCC数据集,结合转移相关基因数据集,探讨HCC患者转移相关基因的预后作用。首先,筛选出222个差异表达的转移相关基因,并对这些差异基因进行GO和KEGG功能注释,发现主要富集在上皮细胞增生,胶原分解代谢和间质发育等过程,这与肝癌具有高度侵袭和转移的特性一致,而KEGG通路富集分析发现主要参与了影响肿瘤细胞侵袭迁移的通路,如细胞因子−细胞因子受体相关作用通路[15],IL-17[16]和Hippo信号通路[17]。同时,通过PPI蛋白质互作网络,筛选出TCGA与ICGC共同的节点基因,并逐步建立了风险预后模型。模型性能评估结果显示TCGA训练集、TCGA验证集以及ICGC外部验证集的分析结果一致。高风险组的患者生存时间和生存状态都明显比低风险组患者的生存时间和生存状态差。各数据集的1年、2年和3年的ROC曲线均大于0.6,表明该模型具有很好的特异性和敏感性。此外,独立预后分析结果表明,该模型的风险评分可独立于其他临床性状,作为HCC患者的独立预后因素。同时,通过临床组织水平的验证,本文发现该模型基因在肝癌组织中的表达水平明显高于癌旁组织,提示其可能作为促癌基因影响肝癌患者的病情进展。基于上述结果,本文发现该转移相关基因预后模型可以为HCC患者的预后提供可靠预测,协助临床个体化治疗方案的制定,并为HCC的基础研究提供潜在的研究靶点。
构成该预后模型的4个转移相关的基因分别是STC2、CDCA8、CTHRC1和HOXD9,其中STC2和HOXD9在HCC中已有研究报道。研究发现,斯钙素-2(STC2)参与了多种癌症的发生和发展,如结直肠癌[18]、乳腺癌[19]、肝癌[20]及头颈部鳞状细胞癌[21]。文献[22]发现STC2可通过AKT-ERK信号通路促进结直肠癌的上皮间质转化,从而影响结直肠癌患者的生存进展。文献[19]发现STC2可通过PKC/claudin-1信号通路抑制乳腺癌细胞的迁移和侵袭,有望成为乳腺癌转移和靶向治疗的生物标志物。细胞分裂周期相关蛋白8(CDCA8)作为癌基因,在多种肿瘤中表达上调[23-25],文献[24]研究发现,过表达CDCA8基因将促进皮肤黑色素瘤的恶性进展,并导致不良预后。文献[25]发现磷酸化的CDCA8明显促进了肺癌细胞的生长,并且CDCA8的过表达与肺癌患者的不良预后密切相关。胶原蛋白的三螺旋重复序列-1(CTHRC1)与肿瘤的发生和转移密切相关,文献[26]发现CTHRC1可通过HIF-1α/CXXR4信号通路促进胃癌的转移;文献[27]表明,CTHRC1可通过与整合素β3-Akt信号通路相互作用促进子宫肌层的侵袭,还可通过上调巨噬细胞中的趋化因子受体CX3CR1的表达促进M2样肿瘤相关巨噬细胞的浸润。同时,CTHRC1可促进宫颈鳞状细胞癌的淋巴结转移[28],促进结直肠癌的EMT转化[29]以及可激活β3/FAK信号通路促进卵巢癌的转移[30]。此外,在文献[14]中,同源框基因9(HOXD9)作为癌基因,在肝细胞癌中高表达,并可直接靶向下游基因ZEB1调控HCC的EMT过程和肿瘤转移。文献[31]也发现miR-205可直接靶向抑制HOXD9表达,从而抑制人脑胶质瘤EMT过程和肿瘤生长。可见,STC2和HOXD9在HCC患者中的作用机制已有深入研究,与本文的发现一致,均能影响HCC患者预后,但暂时缺少关于CDCA8和CTHRC1对HCC患者预后的影响研究。本文发现CDCA8和CTHRC1也参与了HCC患者的预后判断,可作为HCC患者独立预后因素。
-
综上,基于TCGA-HCC数据集和ICGC-HCC数据集,结合转移相关基因,本文构建了一个转移相关基因的预后模型,并验证了该模型的性能。该模型能预测HCC患者的预后,其1年、2年和3年的ROC曲线都显示了其具有很好的特异性和敏感性,独立预后分析也表明该模型能作为HCC患者的独立预后因素,为HCC患者的预后提供判断。但研究尚存在不足,仍需进一步的基础实验来阐明该模型基因在HCC中的作用机制。
Construction and Validation of a Metastasis-Related Prognostic Model for Hepatocellular Carcinoma Patients
doi: 10.12178/1001-0548.2021301
- Received Date: 2021-10-22
- Rev Recd Date: 2021-12-26
- Available Online: 2022-05-23
- Publish Date: 2022-03-25
-
Key words:
- hepatocellular carcinoma /
- metastasis-related genes /
- prognostic model /
- validation
Abstract: Hepatocellular carcinoma is a malignant tumor with high aggressiveness and metastasis. This study aims to construct a predictive model based on metastasis-related genes (MTGs). Cox analysis and Lasso regression analysis were used to build the prognostic model, and the relationship between the risk score of this model and the survival and clinical characteristics of hepatocellular carcinoma (HCC) patients was analyzed. The results of the study constructed a prognostic model containing four MTGs. In the the cancer genome atlas (TCGA) training set, HCC patients in the low-risk group had lower risk scores, fewer deaths and higher 5-year survival (P < 0.01), Cox analysis showed that the model could predict survival of HCC patients independently of other clinical characteristics (P < 0.01), and the AUC values of 1-, 2- and 3-years were all greater than 0.74 in the time-dependent ROC curve prediction analysis. Meanwhile, the validation set also confirmed the above results. Taken together, we constructed a MTGs prognostic model and it was expected to be an independent prognostic factor for prognostic prediction judgment of HCC patients.
| Citation: | LIU Yanqun, XIONG Rong, XIAO Ting, YANG Yan, LIU Kang, FENG Gang, SONG Guiqin. Construction and Validation of a Metastasis-Related Prognostic Model for Hepatocellular Carcinoma Patients[J]. Journal of University of Electronic Science and Technology of China, 2022, 51(2): 184-193. doi: 10.12178/1001-0548.2021301 |

 ISSN
ISSN 
















 DownLoad:
DownLoad: